36 广义可加模型
相比于广义线性模型,广义可加模型可以看作是一种非线性模型,模型中含有非线性的成分。
- 多元适应性(自适应)回归样条 multivariate adaptive regression splines
- Friedman, Jerome H. 1991. Multivariate Adaptive Regression Splines. The Annals of Statistics. 19(1):1–67. https://doi.org/10.1214/aos/1176347963
- earth: Multivariate Adaptive Regression Splines http://www.milbo.users.sonic.net/earth
- Friedman, Jerome H. 2001. Greedy function approximation: A gradient boosting machine. The Annals of Statistics. 29(5):1189–1232. https://doi.org/10.1214/aos/1013203451
- Friedman, Jerome H., Trevor Hastie and Robert Tibshirani. Additive Logistic Regression: A Statistical View of Boosting. The Annals of Statistics. 28(2): 337–374. http://www.jstor.org/stable/2674028
- Flexible Modeling of Alzheimer’s Disease Progression with I-Splines PDF 文档
- Implementation of B-Splines in Stan 网页文档
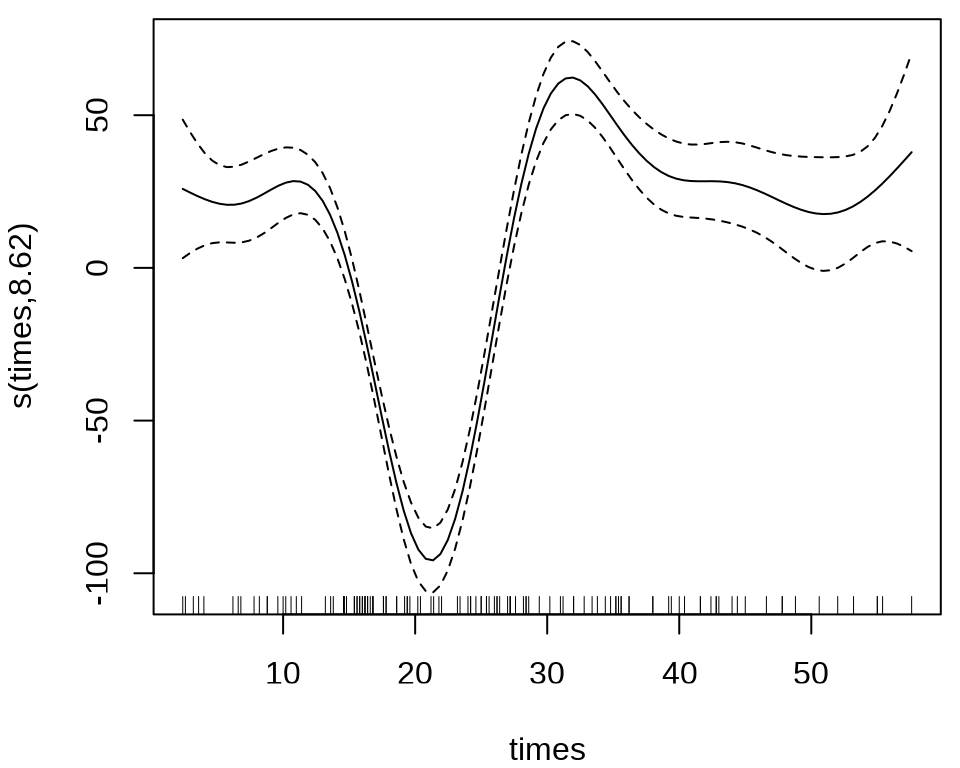
36.1 案例:模拟摩托车事故
36.1.1 mgcv
MASS 包的 mcycle 数据集
data(mcycle, package = "MASS")
library(ggplot2)
ggplot(data = mcycle, aes(x = times, y = accel)) +
geom_point() +
theme_classic() +
labs(x = "时间(ms)", y = "加速度(g)")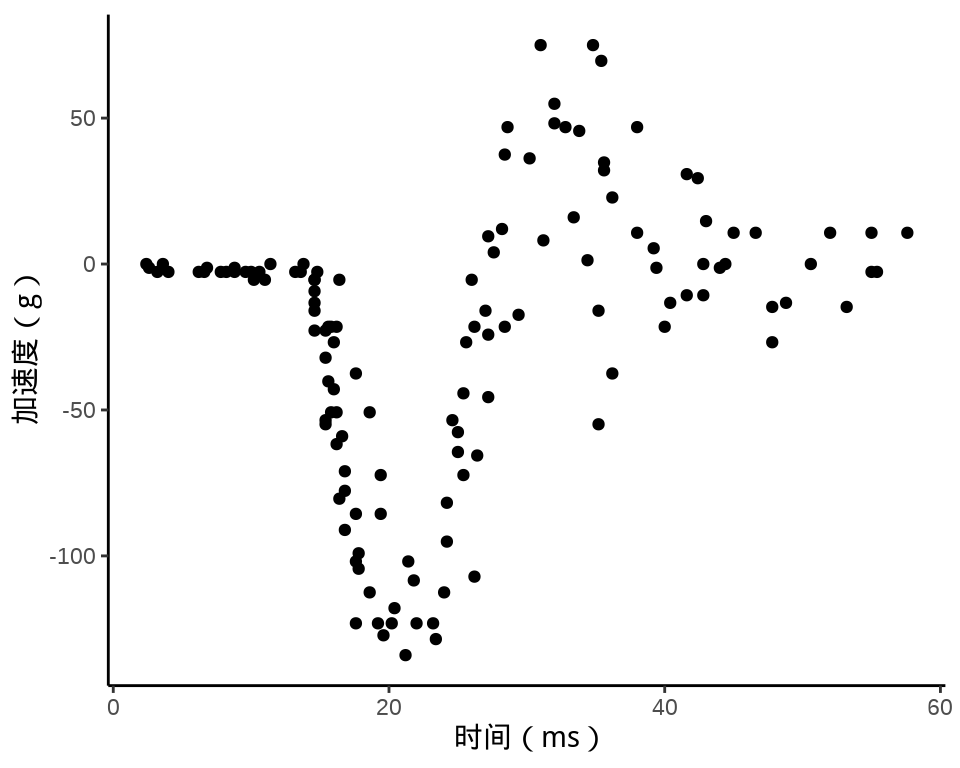
样条回归
#>
#> Family: gaussian
#> Link function: identity
#>
#> Formula:
#> accel ~ s(times)
#>
#> Parametric coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -25.546 1.951 -13.09 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Approximate significance of smooth terms:
#> edf Ref.df F p-value
#> s(times) 8.625 8.958 53.4 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> R-sq.(adj) = 0.783 Deviance explained = 79.7%
#> -REML = 616.14 Scale est. = 506.35 n = 133方差成分
#>
#> Standard deviations and 0.95 confidence intervals:
#>
#> std.dev lower upper
#> s(times) 807.88726 480.66162 1357.88215
#> scale 22.50229 19.85734 25.49954
#>
#> Rank: 2/236.1.2 cmdstanr
36.1.3 rstanarm
rstanarm 可以拟合一般的广义可加(混合)模型。
Model Info:
function: stan_gamm4
family: gaussian [identity]
formula: accel ~ s(times)
algorithm: sampling
sample: 1200 (posterior sample size)
priors: see help('prior_summary')
observations: 133
Estimates:
mean sd 10% 50% 90%
(Intercept) -25.6 2.1 -28.4 -25.5 -23.0
s(times).1 340.4 232.9 61.1 340.8 634.7
s(times).2 -1218.9 243.3 -1529.2 -1218.8 -913.5
s(times).3 -567.8 147.0 -765.2 -567.1 -385.3
s(times).4 -619.8 133.8 -791.1 -617.0 -458.9
s(times).5 -1056.2 85.8 -1162.8 -1055.7 -945.1
s(times).6 -89.2 49.8 -154.4 -89.4 -27.6
s(times).7 -232.2 33.8 -274.7 -232.2 -189.5
s(times).8 17.3 105.8 -121.0 15.5 150.1
s(times).9 4.1 33.1 -25.8 1.0 39.1
sigma 24.7 1.6 22.6 24.6 26.8
smooth_sd[s(times)1] 399.9 59.2 327.6 395.4 479.1
smooth_sd[s(times)2] 25.2 25.4 2.9 17.5 56.6
Fit Diagnostics:
mean sd 10% 50% 90%
mean_PPD -25.5 3.0 -29.3 -25.5 -21.8
The mean_ppd is the sample average posterior predictive distribution of the outcome variable (for details see help('summary.stanreg')).
MCMC diagnostics
mcse Rhat n_eff
(Intercept) 0.1 1.0 1052
s(times).1 7.0 1.0 1103
s(times).2 6.7 1.0 1329
s(times).3 4.4 1.0 1101
s(times).4 3.8 1.0 1230
s(times).5 2.5 1.0 1137
s(times).6 1.5 1.0 1128
s(times).7 1.0 1.0 1062
s(times).8 3.1 1.0 1147
s(times).9 1.0 1.0 1052
sigma 0.0 1.0 1154
smooth_sd[s(times)1] 1.8 1.0 1136
smooth_sd[s(times)2] 0.7 1.0 1157
mean_PPD 0.1 1.0 997
log-posterior 0.1 1.0 1122
For each parameter, mcse is Monte Carlo standard error, n_eff is a crude measure of effective sample size, and Rhat is the potential scale reduction factor on split chains (at convergence Rhat=1).Computed from 1200 by 133 log-likelihood matrix
Estimate SE
elpd_loo -611.0 8.8
p_loo 7.3 1.2
looic 1222.0 17.5
------
Monte Carlo SE of elpd_loo is 0.1.
All Pareto k estimates are good (k < 0.5).
See help('pareto-k-diagnostic') for details.贝叶斯 \(R^2\) 统计量
# 拟合模型
bgam <- brms::brm(accel ~ s(times),
data = mcycle, family = gaussian(), cores = 2, seed = 20232023,
iter = 4000, warmup = 1000, thin = 10, refresh = 0,
control = list(adapt_delta = 0.99)
)
# 模型输出
summary(bgam)
brms::fixef(bgam) # 固定效应
brms::ranef(bgam) # 随机效应
# 边际效应
ms_bgam <- brms::conditional_smooths(bgam) # 平滑效应
plot(ms_bgam)
# R-square
brms::bayes_R2(bgam)
# WAIC
brms::waic(bgam)
# LOOIC
# brms::log_lik(bgam)
brms::loo(bgam)
# 后验预测分布
brms::pp_check(bgam, ndraws = 50)
# 生成代码
brms::stancode(accel ~ s(times), data = mcycle, family = gaussian())36.2 案例:朗格拉普岛核污染
从线性到可加,意味着从线性到非线性,可加模型容纳非线性的成分,比如高斯过程、样条。
36.2.1 mgcv
本节复用 章节 27 朗格拉普岛核污染数据,相关背景不再赘述,下面首先加载数据到 R 环境。
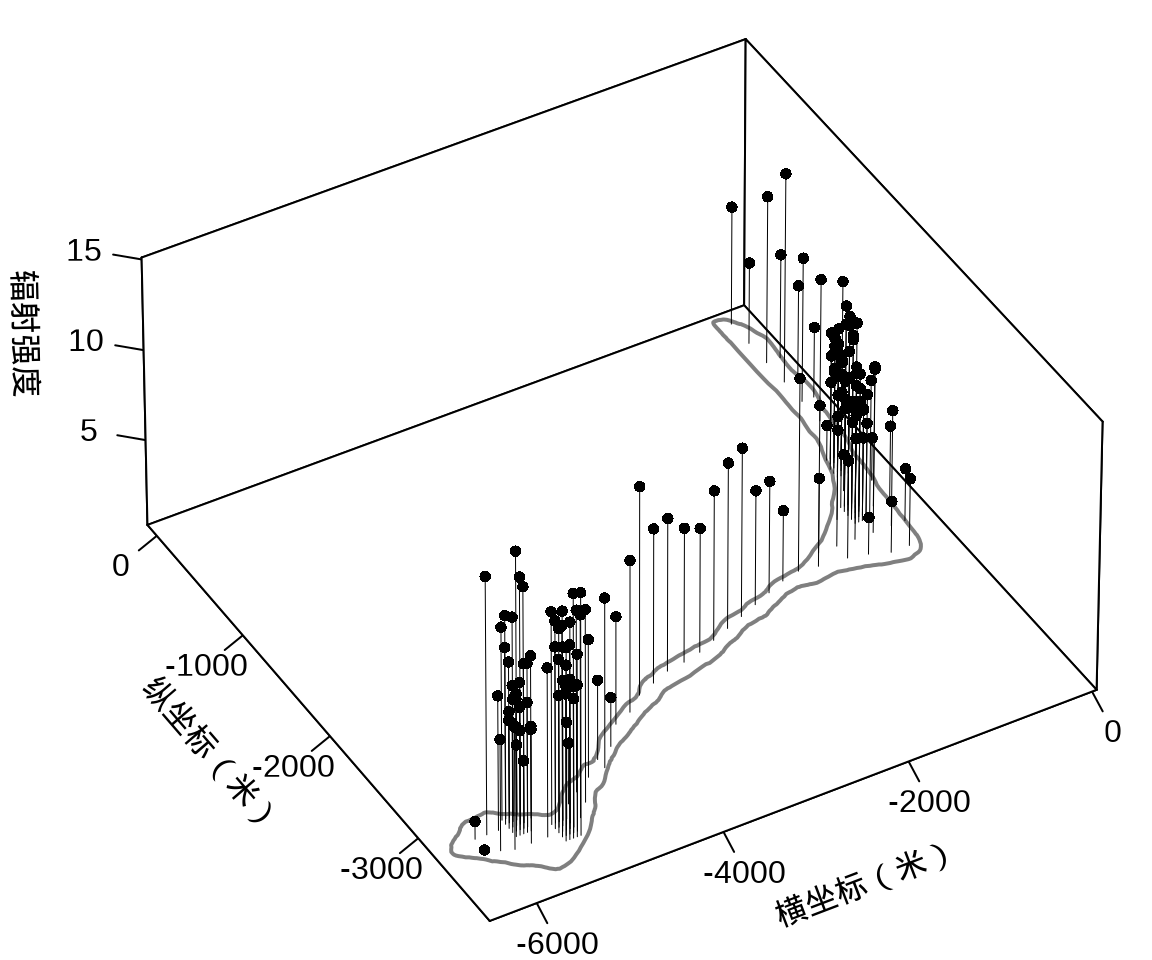
接着,将岛上各采样点的辐射强度展示出来,算是简单回顾一下数据概况。
代码
library(plot3D)
with(rongelap, {
opar <- par(mar = c(.1, 2.5, .1, .1), no.readonly = TRUE)
rongelap_coastline$cZ <- 0
scatter3D(
x = cX, y = cY, z = counts / time,
xlim = c(-6500, 50), ylim = c(-3800, 110),
xlab = "\n横坐标(米)", ylab = "\n纵坐标(米)",
zlab = "\n辐射强度", lwd = 0.5, cex = 0.8,
pch = 16, type = "h", ticktype = "detailed",
phi = 40, theta = -30, r = 50, d = 1,
expand = 0.5, box = TRUE, bty = "b",
colkey = F, col = "black",
panel.first = function(trans) {
XY <- trans3D(
x = rongelap_coastline$cX,
y = rongelap_coastline$cY,
z = rongelap_coastline$cZ,
pmat = trans
)
lines(XY, col = "gray50", lwd = 2)
}
)
rongelap_coastline$cZ <- NULL
on.exit(par(opar), add = TRUE)
})
在这里,从广义可加混合效应模型的角度来对核污染数据建模,空间效应仍然是用高斯过程来表示,响应变量服从带漂移项的泊松分布。采用 mgcv 包 (Wood 2004) 的函数 gam() 拟合模型,其中,含 49 个参数的样条近似高斯过程,高斯过程的核函数为默认的梅隆型。更多详情见 mgcv 包的函数 s() 帮助文档参数的说明,默认值是梅隆型相关函数及默认的范围参数,作者自己定义了一套符号约定。
library(nlme)
library(mgcv)
fit_rongelap_gam <- gam(
counts ~ s(cX, cY, bs = "gp", k = 50), offset = log(time),
data = rongelap, family = poisson(link = "log")
)
# 模型输出
summary(fit_rongelap_gam)#>
#> Family: poisson
#> Link function: log
#>
#> Formula:
#> counts ~ s(cX, cY, bs = "gp", k = 50)
#>
#> Parametric coefficients:
#> Estimate Std. Error z value Pr(>|z|)
#> (Intercept) 1.976815 0.001642 1204 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> Approximate significance of smooth terms:
#> edf Ref.df Chi.sq p-value
#> s(cX,cY) 48.98 49 34030 <2e-16 ***
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#>
#> R-sq.(adj) = 0.876 Deviance explained = 60.7%
#> UBRE = 153.78 Scale est. = 1 n = 157#> s(cX,cY)
#> 2543.376值得一提的是核函数的类型和默认参数的选择,参数 m 接受一个向量, m[1] 取值为 1 至 5,分别代表球型 spherical, 幂指数 power exponential 和梅隆型 Matern with \(\kappa\) = 1.5, 2.5 or 3.5 等 5 种相关/核函数。
接下来,基于岛屿的海岸线数据划分出网格,将格点作为新的预测位置。
#> Linking to GEOS 3.12.0, GDAL 3.7.2, PROJ 9.2.1; sf_use_s2() is TRUElibrary(abind)
library(stars)
# 类型转化
rongelap_sf <- st_as_sf(rongelap, coords = c("cX", "cY"), dim = "XY")
rongelap_coastline_sf <- st_as_sf(rongelap_coastline, coords = c("cX", "cY"), dim = "XY")
rongelap_coastline_sfp <- st_cast(st_combine(st_geometry(rongelap_coastline_sf)), "POLYGON")
# 添加缓冲区
rongelap_coastline_buffer <- st_buffer(rongelap_coastline_sfp, dist = 50)
# 构造带边界约束的网格
rongelap_coastline_grid <- st_make_grid(rongelap_coastline_buffer, n = c(150, 75))
# 将 sfc 类型转化为 sf 类型
rongelap_coastline_grid <- st_as_sf(rongelap_coastline_grid)
rongelap_coastline_buffer <- st_as_sf(rongelap_coastline_buffer)
rongelap_grid <- rongelap_coastline_grid[rongelap_coastline_buffer, op = st_intersects]
# 计算网格中心点坐标
rongelap_grid_centroid <- st_centroid(rongelap_grid)
# 共计 1612 个预测点
rongelap_grid_df <- as.data.frame(st_coordinates(rongelap_grid_centroid))
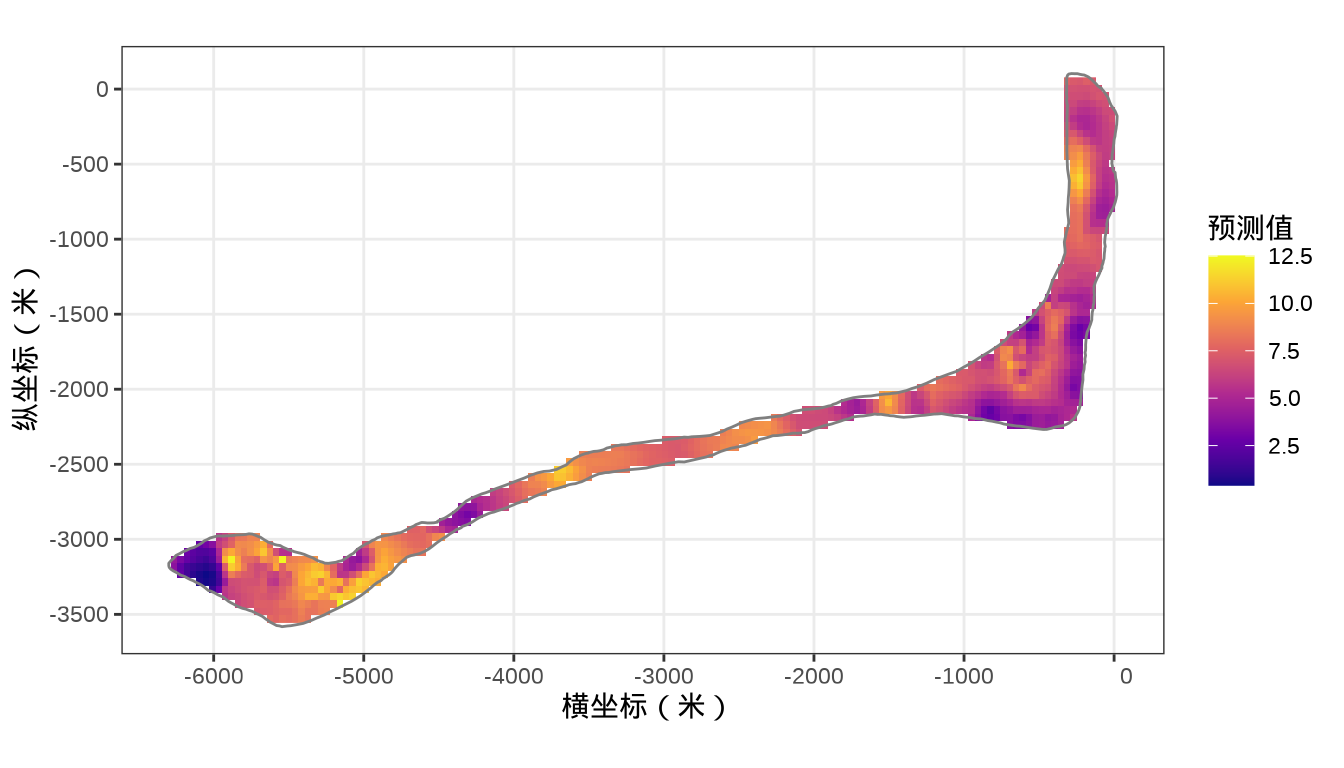
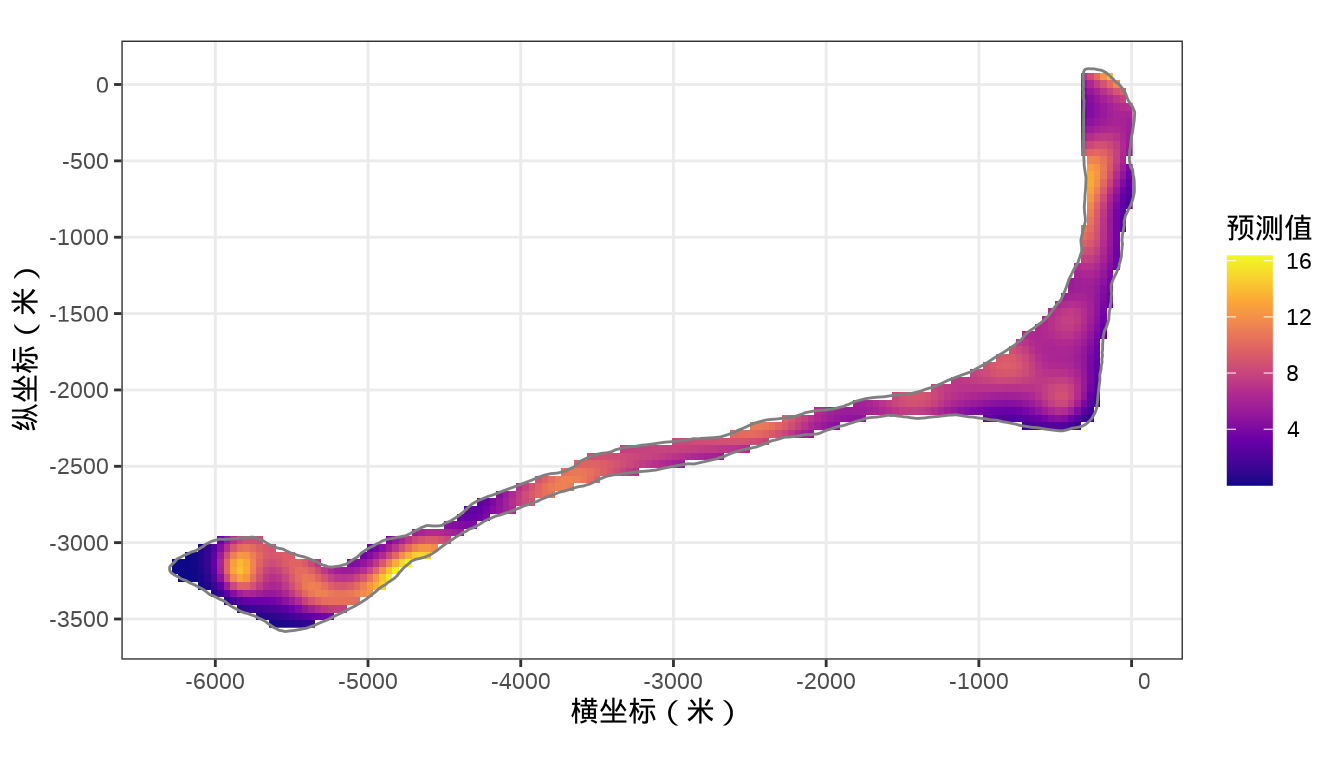
colnames(rongelap_grid_df) <- c("cX", "cY")模型对象 fit_rongelap_gam 在新的格点上预测核辐射强度,接着整理预测结果数据。
# 预测
rongelap_grid_df$ypred <- as.vector(predict(fit_rongelap_gam, newdata = rongelap_grid_df, type = "response"))
# 整理预测数据
rongelap_grid_sf <- st_as_sf(rongelap_grid_df, coords = c("cX", "cY"), dim = "XY")
rongelap_grid_stars <- st_rasterize(rongelap_grid_sf, nx = 150, ny = 75)
rongelap_stars <- st_crop(x = rongelap_grid_stars, y = rongelap_coastline_sfp)最后,将岛上各个格点的核辐射强度绘制出来,给出全岛核辐射强度的空间分布。
代码
36.2.2 cmdstanr
当设置 \(k = 5\) 时,用 5 个基函数来近似高斯过程,编译完成后,采样速度很快,但是结果不可靠,采样过程中的问题很多,见下文。当将横、纵坐标值同时缩小 6000 倍,采样效率并未得到改善。当设置 \(k = 15\) 时,运行时间明显增加,采样过程的诊断结果类似 \(k = 5\) 的情况,还是不可靠。说明 brms 包不适合处理含高斯过程的模型。
Warning messages:
1: There were 4000 transitions after warmup that exceeded the maximum treedepth. Increase max_treedepth above 10. See
https://mc-stan.org/misc/warnings.html#maximum-treedepth-exceeded
2: There were 3 chains where the estimated Bayesian Fraction of Missing Information was low. See
https://mc-stan.org/misc/warnings.html#bfmi-low
3: Examine the pairs() plot to diagnose sampling problems
4: The largest R-hat is 4.4, indicating chains have not mixed.
Running the chains for more iterations may help. See
https://mc-stan.org/misc/warnings.html#r-hat
5: Bulk Effective Samples Size (ESS) is too low, indicating posterior means and medians may be unreliable.
Running the chains for more iterations may help. See
https://mc-stan.org/misc/warnings.html#bulk-ess
6: Tail Effective Samples Size (ESS) is too low, indicating posterior variances and tail quantiles may be unreliable.
Running the chains for more iterations may help. See
https://mc-stan.org/misc/warnings.html#tail-ess 经过对 brms 包生成的模型代码的分析,发现它采用潜变量高斯过程(latent variable GP)模型,这也是采样效率低下的一个关键因素。接下来,分别手动实现低秩和基样条两种方法近似边际高斯过程(marginal likelihood GP)(Rasmussen 和 Williams 2006),用 Stan 编码模型。代码文件分别是rongelap_poisson_lr.stan 和 rongelap_poisson_splines.stan 。
36.2.3 INLA
mgcv 包的函数 ginla() 实现简化版的 Integrated Nested Laplace Approximation, INLA (Rue, Martino, 和 Chopin 2009)。
rongelap_gam <- gam(
counts ~ s(cX, cY, bs = "gp", k = 50), offset = log(time),
data = rongelap, family = poisson(link = "log"), fit = FALSE
)
# 简化版 INLA
fit_rongelap_ginla <- ginla(G = rongelap_gam)
str(fit_rongelap_ginla)#> List of 2
#> $ density: num [1:50, 1:100] 2.49e-01 9.03e-06 3.51e-06 1.97e-06 1.17e-06 ...
#> $ beta : num [1:50, 1:100] 1.97 -676.61 -572.67 4720.77 240.12 ...其中, \(k = 50\) 表示 49 个样条参数,每个参数的分布对应有 100 个采样点,另外,截距项的边际后验概率密度分布如下:
plot(
fit_rongelap_ginla$beta[1, ], fit_rongelap_ginla$density[1, ],
type = "l", xlab = "截距项", ylab = "概率密度"
)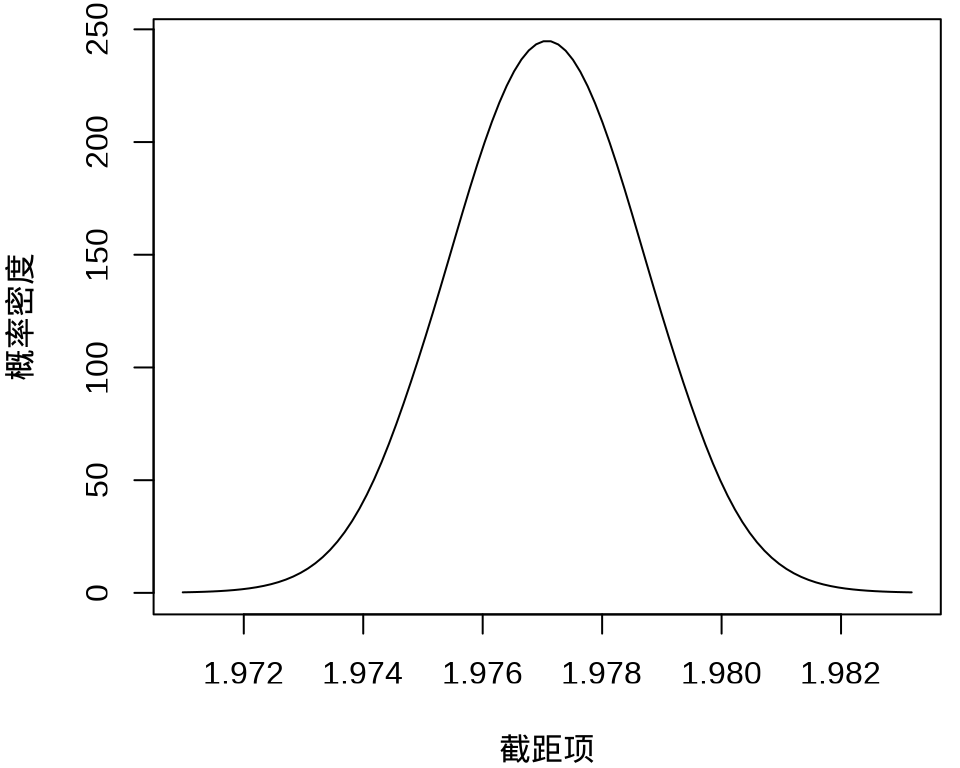
不难看出,截距项在 1.976 至 1.978 之间,50个参数的最大后验估计分别如下:
idx <- apply(fit_rongelap_ginla$density, 1, function(x) x == max(x))
fit_rongelap_ginla$beta[t(idx)]#> [1] 1.977019e+00 -5.124099e+02 5.461183e+03 1.515296e+03 -2.822166e+03
#> [6] -1.598371e+04 -6.417855e+03 1.938122e+02 -4.270878e+03 3.769951e+03
#> [11] -1.002035e+04 1.914717e+03 -9.721572e+03 -3.794461e+04 -1.401549e+04
#> [16] -5.376582e+04 -1.585899e+04 -2.338235e+04 6.239053e+04 -3.574501e+02
#> [21] -4.587927e+04 1.723604e+04 -4.514781e+03 9.184026e-02 3.496526e-01
#> [26] -1.477406e+02 4.585057e+03 9.153647e+03 1.929387e+04 -1.116512e+04
#> [31] -1.166149e+04 8.079451e+02 3.627369e+03 -9.835680e+03 1.357777e+04
#> [36] 1.487742e+04 3.880562e+04 -1.708858e+03 2.775844e+04 2.527415e+04
#> [41] -3.932957e+04 3.548123e+04 -1.116341e+04 1.630910e+04 -9.789381e+02
#> [46] -2.011250e+04 2.699657e+04 -4.744393e+04 2.753347e+04 2.834356e+04接下来,介绍完整版的近似贝叶斯推断方法 INLA — 集成嵌套拉普拉斯近似 (Integrated Nested Laplace Approximations,简称 INLA) (Rue, Martino, 和 Chopin 2009)。根据研究区域的边界构造非凸的内外边界,处理边界效应。
library(INLA)
library(splancs)
# 构造非凸的边界
boundary <- list(
inla.nonconvex.hull(
points = as.matrix(rongelap_coastline[,c("cX", "cY")]),
convex = 100, concave = 150, resolution = 100),
inla.nonconvex.hull(
points = as.matrix(rongelap_coastline[,c("cX", "cY")]),
convex = 200, concave = 200, resolution = 200)
)根据研究区域的情况构造网格,边界内部三角网格最大边长为 300,边界外部最大边长为 600,边界外凸出距离为 100 米。
构建 SPDE,指定自协方差函数为指数型,则 \(\nu = 1/2\) ,因是二维平面,则 \(d = 2\) ,根据 \(\alpha = \nu + d/2\) ,从而 alpha = 3/2 。
生成 SPDE 模型的指标集,也是随机效应部分。
#> s s.group s.repl
#> 691 691 691投影矩阵,三角网格和采样点坐标之间的投影。观测数据 rongelap 和未采样待预测的位置数据 rongelap_grid_df
# 观测位置投影到三角网格上
A <- inla.spde.make.A(mesh = mesh, loc = as.matrix(rongelap[, c("cX", "cY")]) )
# 预测位置投影到三角网格上
coop <- as.matrix(rongelap_grid_df[, c("cX", "cY")])
Ap <- inla.spde.make.A(mesh = mesh, loc = coop)
# 1612 个预测位置
dim(Ap)#> [1] 1612 691准备观测数据和预测位置,构造一个 INLA 可以使用的数据栈 Data Stack。
# 在采样点的位置上估计 estimation stk.e
stk.e <- inla.stack(
tag = "est",
data = list(y = rongelap$counts, E = rongelap$time),
A = list(rep(1, 157), A),
effects = list(data.frame(b0 = 1), s = indexs)
)
# 在新生成的位置上预测 prediction stk.p
stk.p <- inla.stack(
tag = "pred",
data = list(y = NA, E = NA),
A = list(rep(1, 1612), Ap),
effects = list(data.frame(b0 = 1), s = indexs)
)
# 合并数据 stk.full has stk.e and stk.p
stk.full <- inla.stack(stk.e, stk.p)指定响应变量与漂移项、联系函数、模型公式。
# 模型拟合
res <- inla(formula = y ~ 0 + b0 + f(s, model = spde),
data = inla.stack.data(stk.full),
E = E, # E 已知漂移项
control.family = list(link = "log"),
control.predictor = list(
compute = TRUE,
link = 1, # 与 control.family 联系函数相同
A = inla.stack.A(stk.full)
),
control.compute = list(
cpo = TRUE,
waic = TRUE, # WAIC 统计量 通用信息准则
dic = TRUE # DIC 统计量 偏差信息准则
),
family = "poisson"
)
# 模型输出
summary(res)#>
#> Call:
#> c("inla.core(formula = formula, family = family, contrasts = contrasts,
#> ", " data = data, quantiles = quantiles, E = E, offset = offset, ", "
#> scale = scale, weights = weights, Ntrials = Ntrials, strata = strata,
#> ", " lp.scale = lp.scale, link.covariates = link.covariates, verbose =
#> verbose, ", " lincomb = lincomb, selection = selection, control.compute
#> = control.compute, ", " control.predictor = control.predictor,
#> control.family = control.family, ", " control.inla = control.inla,
#> control.fixed = control.fixed, ", " control.mode = control.mode,
#> control.expert = control.expert, ", " control.hazard = control.hazard,
#> control.lincomb = control.lincomb, ", " control.update =
#> control.update, control.lp.scale = control.lp.scale, ", "
#> control.pardiso = control.pardiso, only.hyperparam = only.hyperparam,
#> ", " inla.call = inla.call, inla.arg = inla.arg, num.threads =
#> num.threads, ", " keep = keep, working.directory = working.directory,
#> silent = silent, ", " inla.mode = inla.mode, safe = FALSE, debug =
#> debug, .parent.frame = .parent.frame)" )
#> Time used:
#> Pre = 0.882, Running = 1.7, Post = 0.0791, Total = 2.66
#> Fixed effects:
#> mean sd 0.025quant 0.5quant 0.975quant mode kld
#> b0 1.828 0.061 1.706 1.828 1.948 1.828 0
#>
#> Random effects:
#> Name Model
#> s SPDE2 model
#>
#> Model hyperparameters:
#> mean sd 0.025quant 0.5quant 0.975quant mode
#> Theta1 for s 2.00 0.063 1.88 2.00 2.12 2.00
#> Theta2 for s -4.85 0.130 -5.11 -4.85 -4.59 -4.85
#>
#> Deviance Information Criterion (DIC) ...............: 1834.56
#> Deviance Information Criterion (DIC, saturated) ....: 314.90
#> Effective number of parameters .....................: 156.46
#>
#> Watanabe-Akaike information criterion (WAIC) ...: 1789.31
#> Effective number of parameters .................: 80.06
#>
#> Marginal log-Likelihood: -1331.95
#> CPO, PIT is computed
#> Posterior summaries for the linear predictor and the fitted values are computed
#> (Posterior marginals needs also 'control.compute=list(return.marginals.predictor=TRUE)')kld表示 Kullback-Leibler divergence (KLD) 它的值描述标准高斯分布与 Simplified Laplace Approximation 之间的差别,值越小越表示拉普拉斯的近似效果好。DIC 和 WAIC 指标都是评估模型预测表现的。另外,还有两个量计算出来了,但是没有显示,分别是 CPO 和 PIT 。CPO 表示 Conditional Predictive Ordinate (CPO),PIT 表示 Probability Integral Transforms (PIT) 。
固定效应(截距)和超参数部分
#> mean sd 0.025quant 0.5quant 0.975quant mode kld
#> b0 1.828057 0.06149195 1.706415 1.828314 1.948234 1.828309 1.782788e-08#> mean sd 0.025quant 0.5quant 0.975quant mode
#> Theta1 for s 2.000465 0.06252247 1.876613 2.000728 2.122786 2.001821
#> Theta2 for s -4.851577 0.12998555 -5.106820 -4.851802 -4.595020 -4.852739提取预测数据,并整理数据。
# 预测值对应的指标集合
index <- inla.stack.index(stk.full, tag = "pred")$data
# 提取预测结果,后验均值
# pred_mean <- res$summary.fitted.values[index, "mean"]
# 95% 预测下限
# pred_ll <- res$summary.fitted.values[index, "0.025quant"]
# 95% 预测上限
# pred_ul <- res$summary.fitted.values[index, "0.975quant"]
# 整理数据
rongelap_grid_df$ypred <- res$summary.fitted.values[index, "mean"]
# 预测值数据
rongelap_grid_sf <- st_as_sf(rongelap_grid_df, coords = c("cX", "cY"), dim = "XY")
rongelap_grid_stars <- st_rasterize(rongelap_grid_sf, nx = 150, ny = 75)
rongelap_stars <- st_crop(x = rongelap_grid_stars, y = rongelap_coastline_sfp)最后,类似之前 mgcv 建模的最后一步,将 INLA 的预测结果绘制出来。